Evaluation beyond accuracy
October 21, 2025
TLDR: GraphFLA enables efficient characterization of landscape features of arbitrary sequence-fitness landscapes, thereby augments the performance evaluation of genomic models
Abstract #
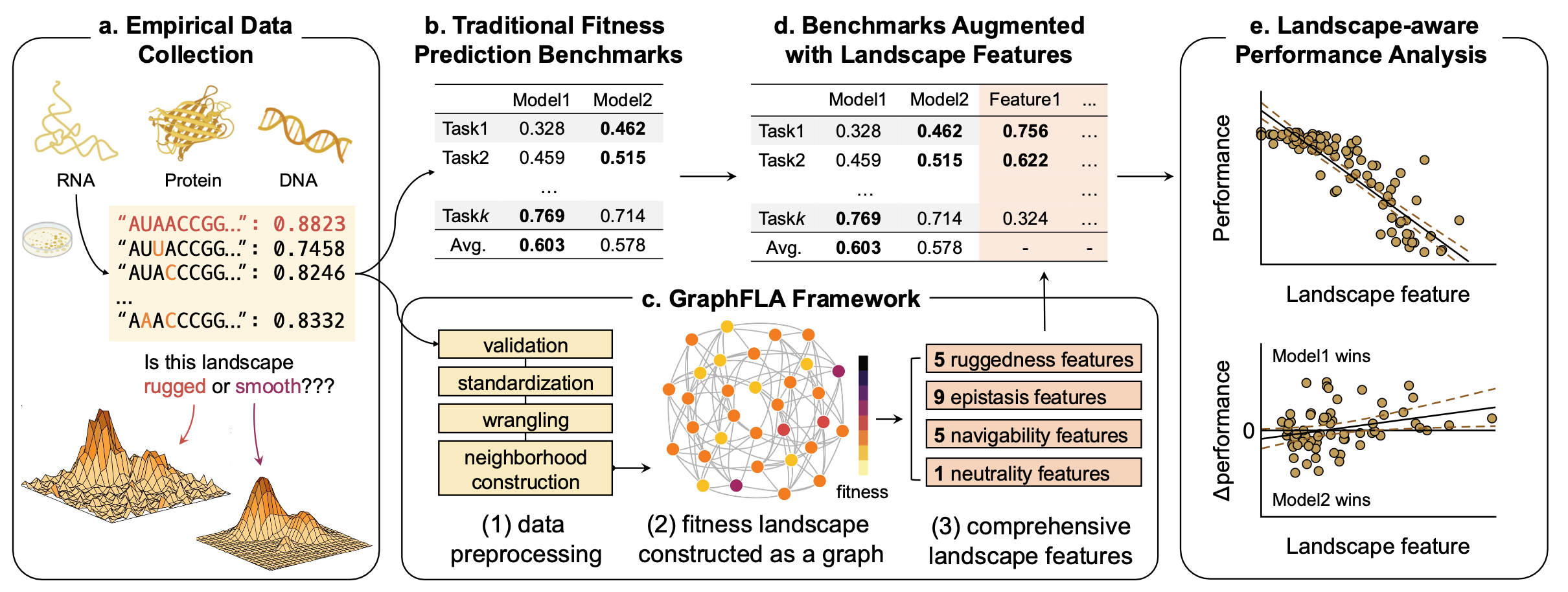
Machine learning models increasingly map biological sequence-fitness landscapes across various modalities to predict mutational effects. Effective evaluation of these models requires benchmarks derived from empirical data. However, current benchmarks often lack detailed topographical information regarding the underlying fitness landscape, which hampers interpretation and comparison of model performance. Here, we introduce GraphFLA, a Python framework that constructs and analyzes fitness landscapes from diverse modalities (DNA, RNA, protein, and beyond), accommodating datasets up to millions of mutants. GraphFLA calculates 20 biologically relevant features that characterize 4 fundamental aspects of landscape topography.
By applying GraphFLA to over 5,300 landscapes from ProteinGym, RNAGym, and UniProbe, we demonstrate its utility in interpreting and comparing the performance of dozens of fitness prediction models, highlighting factors influencing model accuracy.
Background and Scope #
This work considers a fundamental problem in genomics: learning the sequence-to-fitness map (a.k.a. the fitness landscape). Across all three modalities of the central dogma (DNA, RNA, protein), numerous predictive models have been developed for this goal, and various benchmark suites have been established to assess their performance (e.g., ProteinGym, RNAGym).
The Statu Quo #

Existing benchmarks often feature dozens to hundreds of tasks (e.g., the famous ProteinGym benchmark has 217 DMS substitution tasks), wherein each is derived from a specific fitness landscape (e.g., the famous GB1 landscape in protein engineering). While this rich of tasks allows for a more comprehensive evaluation of genomic models, current interpretation of the benchmark results mainly rely on averaged scores across all tasks, which can often lead to misleading results as there is no model that can dominate on all tasks (i.e., the famous “no free lunch” theorem).
In particular, this leads to two unresolved questions:
- Q1: Why did one model perform well on one set of tasks but poorly on another?
- Q2: Why did on model outperform baseline on one task, but not on the other?
Anwering these questions require effective meta-features to characterize the landscape topography of each task, which is unfortunately not available in all existing genomic benchmarks.
The GraphFLA Framework #

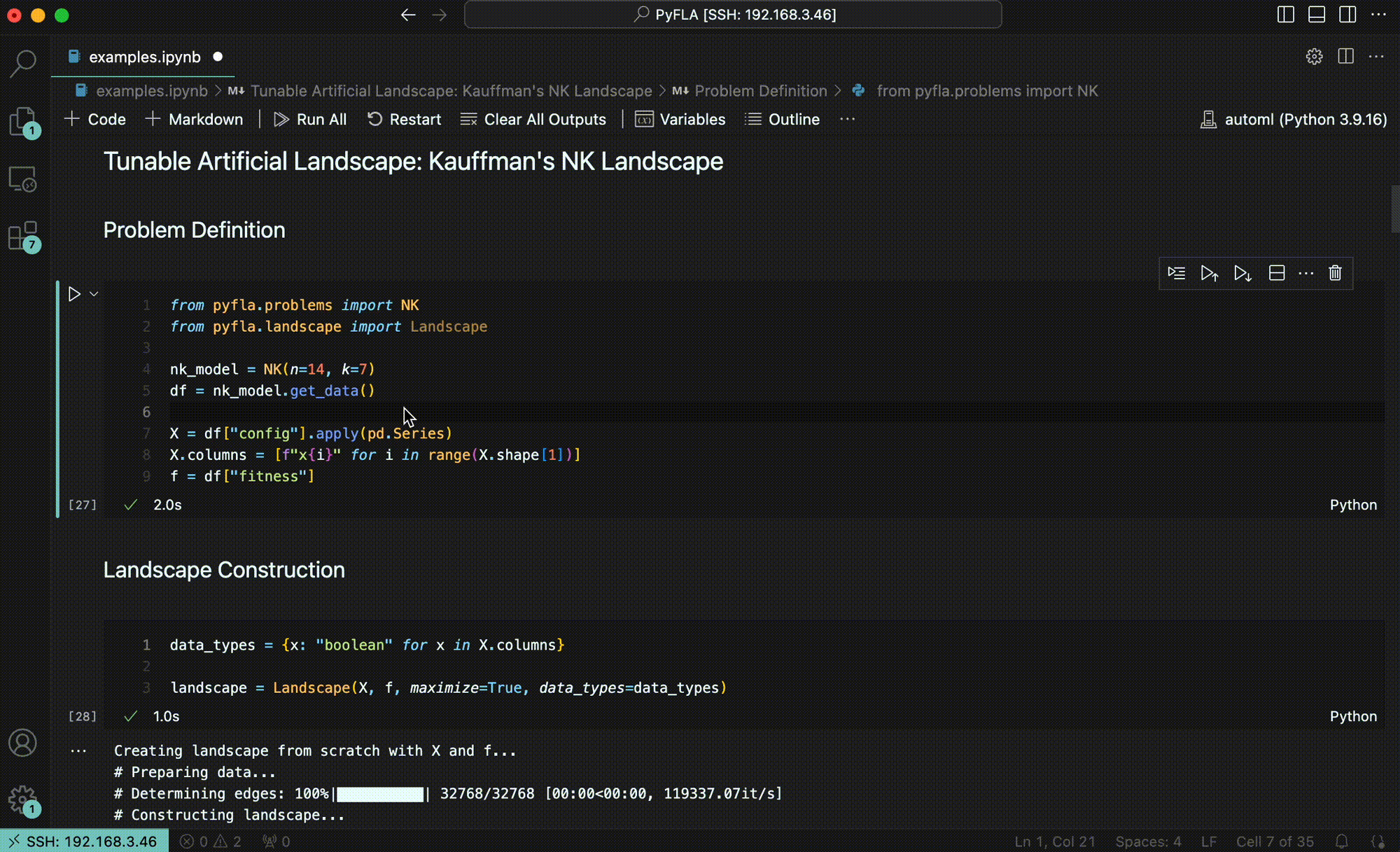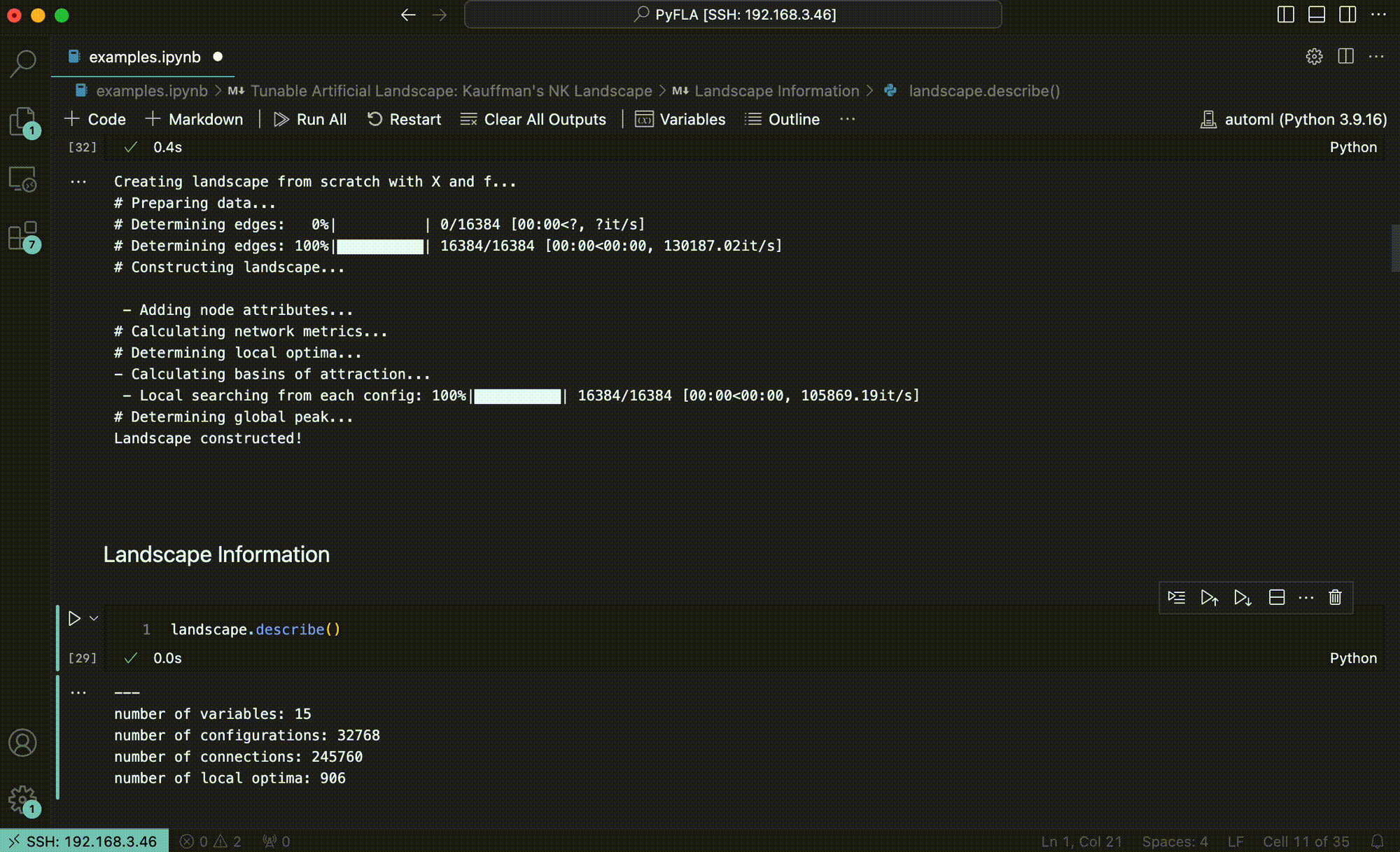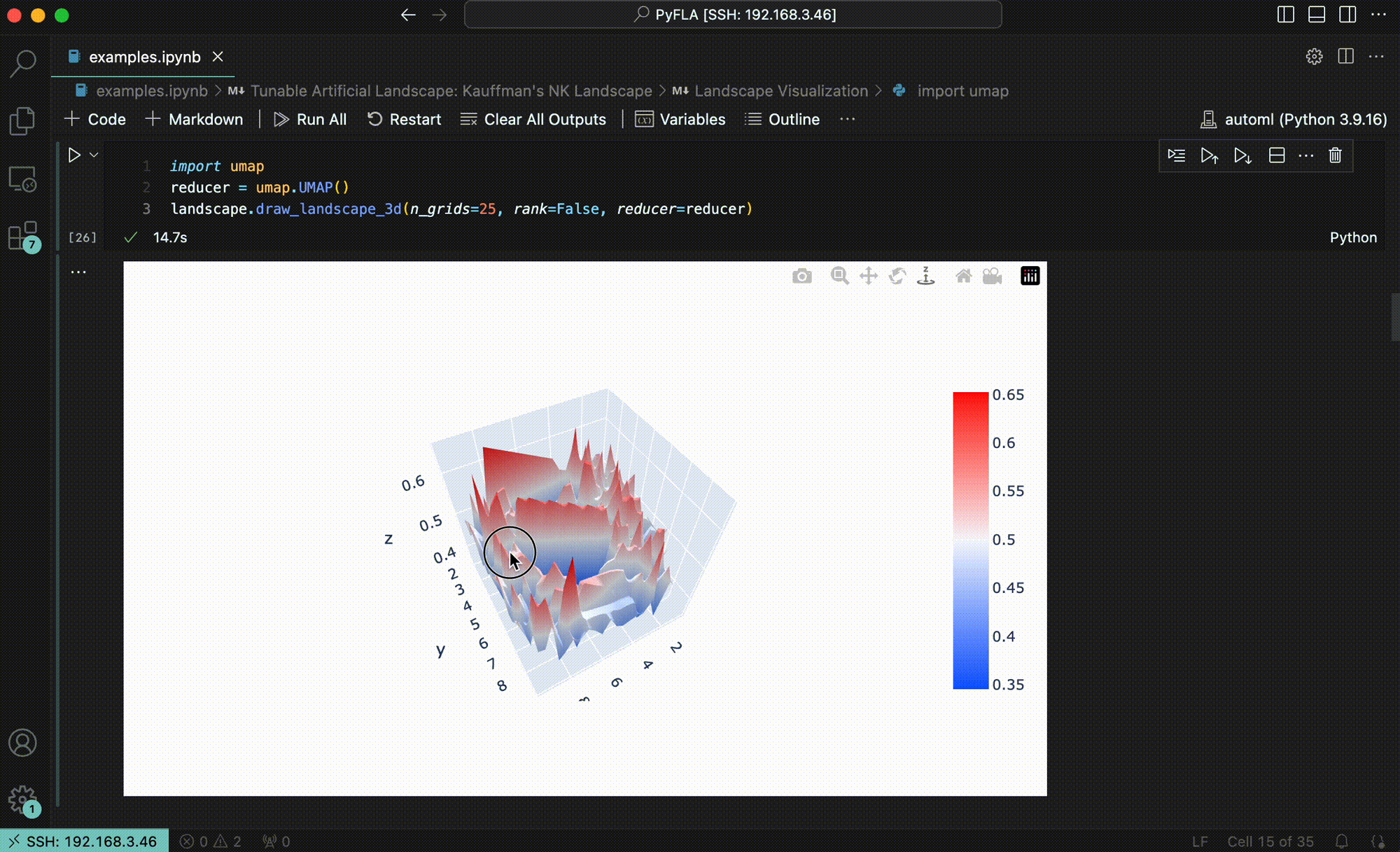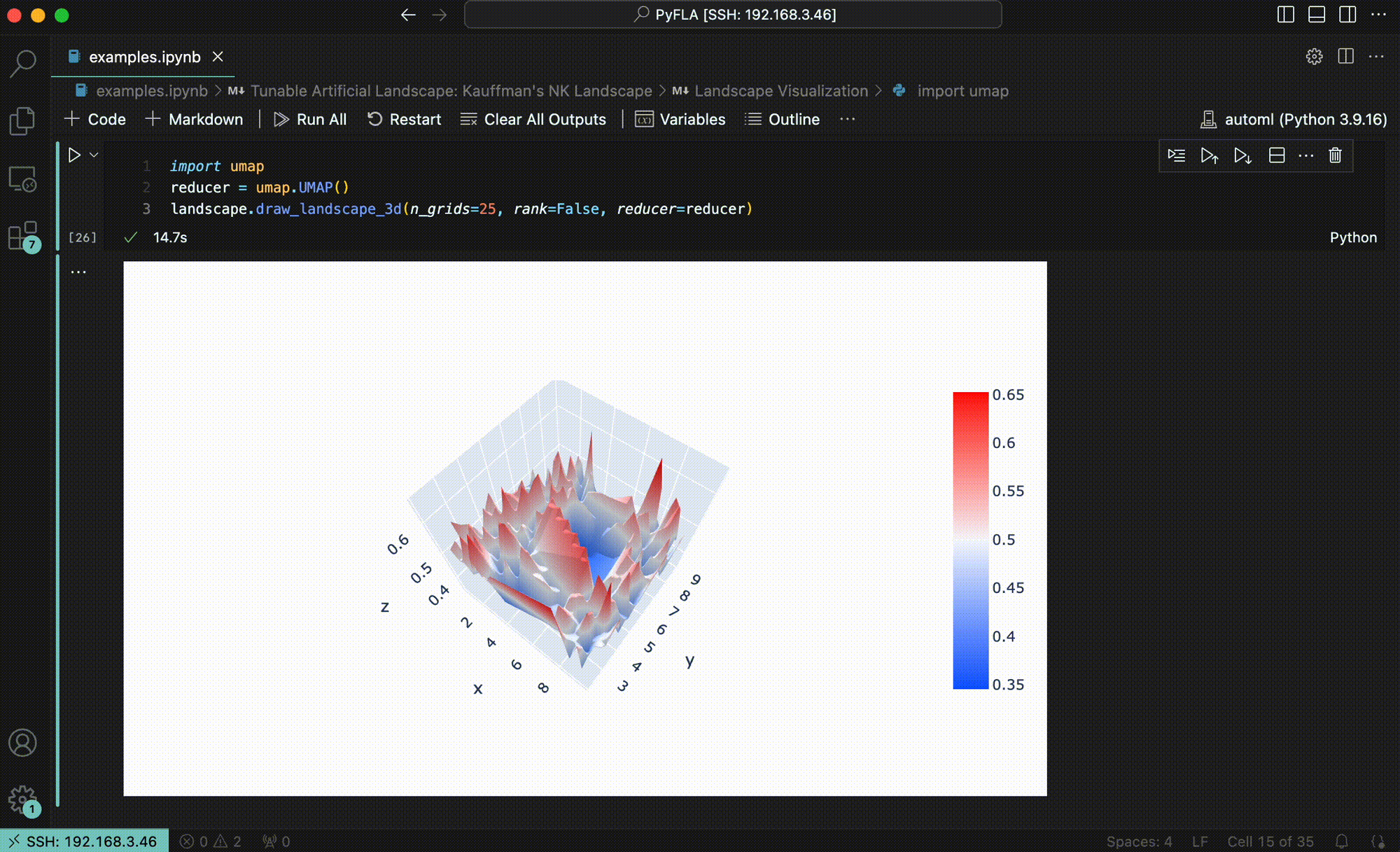
To address this gap, we developed GraphFLA, a modular, efficient Python framework for calculating fitness landscape features for arbitrary sequence-to-function landsacpes. It takes the sequence-to-function data as the input, and automatically construct a directed, attributed graph to represent the underlying landscape. Once constructed, it can then calculate a holistic set of 20 landsacpe features derived from evolutionary biology with just a few lines of code. These features cover 4 essential aspects of landscape topography:
- Ruggedness
- Epistasis
- Navigability
- Neutrality
Using GraphFLA to Enchance Model Evaluation #
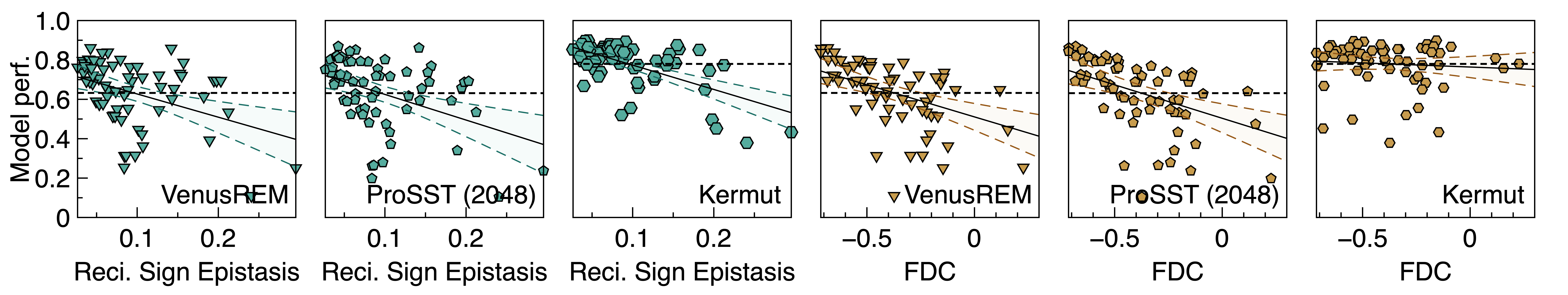
With GraphFLA’s landscape features, more granular interpretations of the performance results from existing benchmarks can be obtained. For example, for Q1, we found that existing genomic models struggle at landscapes that are more rugged, epistatic, neutral, while less navigable.
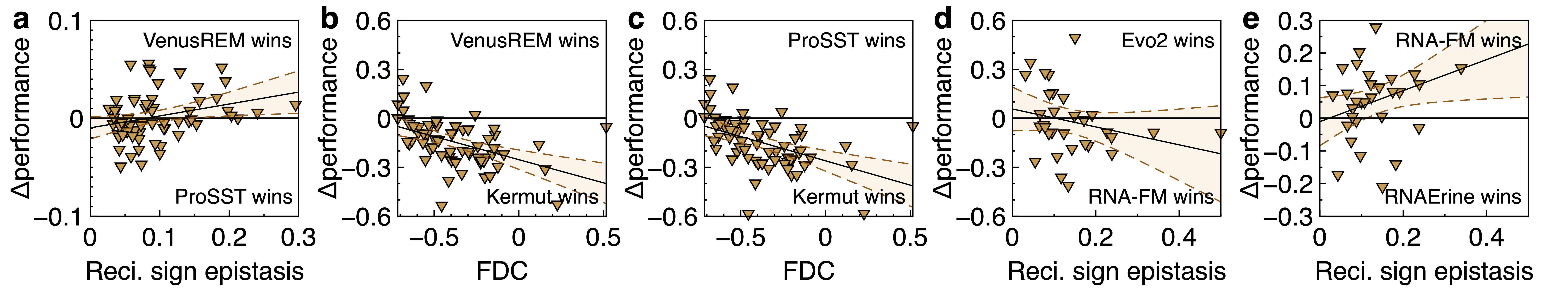
For Q2, we found that the performance gaps between different models can vary with landscape features. For example, on ProteinGym, ProSST tends to perform well on benign landscapes with little reciprocal sign epistasis, while VenusREM leads on more rugged ones.
Outlook #
With this work, we hope to raise the awareness of the community on adopting more granular analysis on performance benchmarks of genomic models, instead of simply relying on averaged scores. GraphFLA serves as an effective tool for this purpose, as we have shown that by augmenting existing benchmarks with landscape features, we can already obtain various interesting insights that were unachivable before.
BibTeX #
@article{zhou2025autovla,
title = {Augmenting Biological Fitness Prediction Benchmarks with Landscapes Features from GraphFLA},
author = {Mingyu Huang, Shasha Zhou, Ke Li},
booktitle = {{NeurIPS}'25: Proc. of the 39th Conference on Neural Information Processing Systems},
year = {2025}
}